Our lab has been sending samples off to a lab out of state for 454 sequencing of amplicon pools for almost a year now. We have so far just used this to assess fungal diversity (ITS region), but in theory could come up with all sorts of applications. Other labs at NAU have been doing the same thing, but look at various targets to assess various questions of diversity (bacterial, archaeal, fungal, functional gene diversity etc). Another tool I routinely use is tailed fluorescent primers for labeling microsatellites I amplify from my field studies (seeSchuelke, M. (2000). An economic method for the fluorescent labeling of PCR fragments. Nature Biotechnology, 18, 233-234). Not too long ago, I was thinking tailed primers would be useful for amplifying samples for 454 samples, but it is very expensive to just test this idea out. Instead, I used it successfully for doing tRFLPs for QC prior to sending samples off for 454 sequencing. Later I realized this was discussed in the GS Junior Guide for Experimental Design, though some details and crucially, a discussion of the data produced, was missing.
So I was excited to see a couple of recent publications:
Bybee, S. M., Bracken-Grissom, H., Haynes, B. D., Hermansen, R. a, Byers, R. L., Clement, M. J., Udall, J. a, et al. (2011). Targeted amplicon sequencing (TAS): A scalable next-gen approach to multi-locus, multi-taxa phylogenetics. Genome biology and evolution, 3, 1312-1323.
Daigle, D., Simen, B., & Pochart, P. (2011). High-Throughput Sequencing of PCR Products Tagged with Universal Primers Using 454 Life Sciences Systems. Current Protocols in Molecular Biology, 96, 7.5.1-7.5.14.
The authors carefully go through the steps involved in using tailed primers and detail their results. So in order to maximize the monetary power of all labs doing this here at NAU, I proposed to try using a set of tailed primer to barcode a sample for 454 amplicon sequencing. I did not spend a lot of time, but things did not go as smoothly as I had hoped.
Here are the primers I used:
M13f-ITS1F 5'-CGCCAGGGTTTTCCCAGTCACGACCTTGGTCATTTAGAGGAAGTAA-3'
M13r-ITS4 5'-TCACACAGGAAACAGCTATGACTCCTCCGCTTATTGATATGC-3'
LibAf-MID01-M13f 5'-CGTATCGCCTCCCTCGCGCCATCAGACGAGTGCGTCGCCAGGGTTTTCCCAGTCACGAC-3'
LibAr-MID01-M13r
5'-CTATGCGCCTTGCCAGCCCGCTCAGACGAGTGCGTTCACACAGGAAACAGCTATGAC-3'
LibAf 5'-CGTATCGCCTCCCTCGCGCCATCAG-3'
LibAr 5'-CTATGCGCCTTGCCAGCCCGCTCAG-3'
The M13-ITS primers have the ITS primer 3' and the M13 sequence 5'. The LibA-MID-M13 primers have LibA sequence 5', MID01 in the middle, and M13 sequence 3'.
First amplification (20uL reactions, 0.01U/uL Phusion polymerase, MgCl2 at 2.5mM, M13-ITS primers at 200nM; 30 cycles of 90C 30s, 57C 30s, 72C 1min)
There are 4 samples in the gel (2uL/well). First four wells are each sample at 1X DNA concentration, 2nd four DNA at 1/20X. Ladder is (bp) 2000, 800, 400, 200, 100. Note the significant artifact around 100bp.
I column purified the 1/20X products with Epoch Life Science columns (http://www.epochlifescience.com/Product/SpinColumn/minispin.aspx) and quantified with OD260. All purified products were around 40 ng/uL.
Here is a gel of the column purified products (10uL/well):
So I co-purified my 100bp artifact, but I continued nonetheless. To try to reduce the amplification of the non-specific product, I added 2.5uL each purified product to 1000uL 10mM Tris-Cl pH 8.8. I amplified under same conditions both the raw column purified product and the diluted product using LibA-MID01-M13 primers this time:
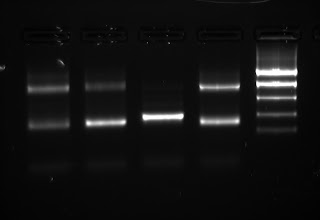
First 4 samples are using full-strength column purified DNA, second 4 samples are using the dilution. The ITS product looks really weak now, and the 100bp artifact has dominated my PCR. No bueno!
I reran the raw M13-ITS products on a gel and cut out the ITS bands this time. I was generous in the size of gel slices in case there were products I couldn't see. Briefly, I put the gel slice in a 1.5mL tube, added 3 2mm steel beads, and beat the gel in the GenoGrinder for 15s at about 28hz. I spun the gel down briefly, added 200uL Tris-Cl, vortexed, and placed in 65C block for 5 min. I spun 1 min at 15,000rpm, and pipeted off 100uL of supernatant. Gel-purified products extruded!
I then redid the LibA tagging using the LibA-MID01-M13 primers on my gel-purified samples (full strength only):
Each 2 wells are 8uL column purified M13-ITS product, then 2uL LibA-tagged product from the last PCR round. You can see the gel purification made a huge difference in maintaining specificity. You can also see the subtle gel shift as the LibA tagging incorporates another 50bp or so. Still, some unwated product remains, particularly now at about 180bp.
So I thought I could change PCR conditions and improve things. I cut out the excess MgCl2 so that it was back to 1.5mM and used the following PCR conditions: 30 cycles of 90C 30s, 63C 2 min, 72C 15s. Intial PCR (M13-ITS primers) yielded OK results:
The top comb is from another project, but the bottom comb are the same four samples in a PCR serial dilution (2uL/well). First 4 samples 1X DNA; 2nd 4, 1/10X DNA; 3rd 4, 1/100X DNA. I thought perhaps the column cleanup was unnecessary and perhaps carryover primer was causing my 180bp non-specific product in the LibA tailing step so I thought I would use Exonuclease I to clean up the samples this time (just the 1X and 1/10X products).
Using the same altered PCR conditions, I did the LibA tailing step:
(First 4 wells, 1X DNA template, 2nd 4 wells, 1/10X template, 9th well, no DNA template). That pesky 180bp artifact is still present, so ExoI didn't help in this case. That, plus the negative control this time mean that the artifact is definitely double-stranded primer-dimer of some sort. Now I reran the first PCR of this series (M13-ITS primers) on a gel and did another gel extraction (1X and 1/10X products):
You can see the 180bp product is pretty strong still and the ITS product is present, but weak. I attribute this to the fact that the initial ITS product with this method was pretty weak. I think it could be partially resolved by adding a few more cycles to the PCR and then doing the gel extraction. Having your product of interest in molar excess seems crucial to this process.
Chatting with a colleague, he was bothered by the number of PCR reactions needed to generate a 454 amplicon library this way. I agreed that it would unduly bias your pool so that relative abundances would be almost meaningless and then I thought of another solution. Put all primers in the same mix, and just do a single amplification! I didn't have time to redo the initial amplification, even though I probably should have, but I thought I could use the last gel-purified products which used the M13-ITS primers:
These are products just from the 1/10X gel-purified products as template (2uL/well). I included LibA primers (no tail) at 200nM each as drivers of the desired products along with LibA-MID01-M13 primers at 20nM each to get the reaction going. As you can see, the 180bp product is quite present, but the ITS products came out much better this time. Perhaps using a stronger initial M13-ITS product or even including M13-ITS in the whole mix would yield a cleaner amplicon pool. For now I am out of time for this, and these products are not sequenceable. We will order a set of barcoded primers specific to the loci we are interested in a gasket our 454 plates until something better comes along or we solve the non-specific product issue. The Bybee et al (2011) reference stated that such non-specific products used up a third of the reads on their plates. We can't have that!







http://www.ncbi.nlm.nih.gov/pubmed/21947461
ReplyDeletehttp://aem.asm.org/content/77/21/7846.abstract
http://www.nature.com/ismej/journal/v1/n4/full/ismej200753a.html
These reports showed that doing sequential labeling cycles (overlapping PCRs for the adaptors)is actually indtroducing less biases than single PCR with long primers. From 1/10 of the amplicon, 5 to 10 cycles in the fresh reaction is enough.
Thanks Howard. Those were interesting references, especially the AEM paper. It leaves me thinking I needn't worry so much about the barcode sequences themselves, though running through barcrawl (http://www.ncbi.nlm.nih.gov/pubmed/19874596) can't hurt, and more about doing reduced PCR cycles in library preps. I think I was on the right track, but happy these guys worked it all out already.
DeleteI wonder if having 2 different annealing temperatures helps. The first 5-10 steps will be lower to accommodate just the locus specific part and the higher temp (72C) will be to incorporate the whole primers.
ReplyDeleteI know this isn't at all timely, but I do have an update on this.
ReplyDeleteIn October, we acquired our very own Miseq so we no longer are interested in 454 sequencing. The Berry et al paper suggested by Howard above was most interesting to me. I have been using this approach for producing amplicons for several months now and it definitely cleans up the primer problem. Also, I have been playing with mock communities recently trying to find a way to maintain community representation throughout all the sample prep so that the results can be interpreted in a quantitative fashion. Some people do this already, but extreme caution must be taken when doing so. The main reason for this is that overamplified libraries exhibit skewed representations of the various products (whether for amplicon sequencing, genomic, or otherwise), so one cannot simply say that taxon A is present in twice the amount as taxon B according to some 454 data and expect people to believe them. So to produce strong products for sequencing but to avoid bias, I have adopted an unusual PCR strategy that couples long anneal/extension time with low cycle number. Using 0.01U/uL Phusion HotStart II polymerase I am able to produce observable product on a gel (2uL) after just 10 cycles. The protocol goes like this:
1) 95C 2min
2) 95C 30s
3) 55C 30s
4) 60C 4min
5) goto 2, 9 times
Bring your MgCl2 to 3.0mM, and for the locus I have been using, 515/806 (bacterial/archaeal 16S v4), I bring the primers to 1mM in the reaction since each has several degenerate positions.
Finally, I no longer favor column preps for anything!! Bead preps are where it's at. I've been using size selection bead solution from Epoch Life Science (http://www.epochlifescience.com) for a while now which are still spendy, but offer savings over AMpure, and they work just fine. A recipe for your own bead solution was published in Genome Research and detailed here (http://www.molecularecologist.com/2012/01/a-penny-for-your-method-ampure-substitute/). Given the cost savings for homemade bead solution, I now only want to use beads if I am faced with any sort of cleanup step. Since they rely on PEG precipitation to facilitate adsorption of DNA to the beads, these methods are also excellent at removing low molecular weight contamination and one can "tune" the desired size for a given cleanup. Since it requires more PEG to precipitate smaller DNA fragments, this means using them at higher volume to sample ratios could selectively precipitate smaller products (useful if you wanted to cleanup a bigdye reaction, for instance).
I was just looking over some things and I noticed I made an error in the previous comment. Primers are being used at 1uM to compensate for degeneracy. I do this for ITS2 and 18S primers for fungal amplicons now as well.
ReplyDelete